Washington Road
Princeton, NJ 08544
Jillian Gullo
217 Schultz Laboratory
jg8071[at]princeton.edu
(609)-258-5028
 Much like human beings, microbes often live in diverse communities interacting with both collaborators and competitors. Small molecule natural products mediate a significant portion of these interactions. As expected, the more complex a microbial community is, the richer its small molecule chemical arsenal becomes.
Much like human beings, microbes often live in diverse communities interacting with both collaborators and competitors. Small molecule natural products mediate a significant portion of these interactions. As expected, the more complex a microbial community is, the richer its small molecule chemical arsenal becomes.
In addition to the de novo production of small molecules, the human gut microbiome encodes a large repertoire of biochemical enzymes that can metabolize exogenous small molecules – whether they are derived from the human host, or from dietary or therapeutic sources. This concept has been explored for decades by using single isolates of the human microbiome against a common set of human-, drug-, or diet-derived chemicals, but has not been explored systematically. We aim to expand this area of research by developing high-throughout, unbiased approaches for defining the space of biochemical transformations that can be exerted by the collective human gut microbiome against a wide variety of chemicals, and the pharmacological consequences of these transformations.

In the case of marine organisms (e.g., sponges, tunicates, mollusks, algae) and their symbionts, small-molecule-mediated interactions can provide the host with indispensable means of chemical defense, allowing it to survive in a predator-rich environment. We characterize these interactions by first determining the true microbial producers of the defensive molecules and the molecular mechanisms of their production, then we study the evolution of the symbiosis in light of the production of these molecules, and finally, we study the ecological consequences of their production for all interacting partners.
With respect to the human body and its microbial inhabitants (the human microbiome), small-molecule-mediated interactions can define the difference between commensals and pathogens, and thus between health and disease states. We are interested in studying these interactions at three main levels: a) identifying the chemical repertoire of the human microbiome; b) characterizing the biological activities and functional roles of the identified molecules in mediating microbiome-host interactions in health and disease states; and finally c) using this knowledge to aid the diagnosis and treatment of human diseases via microbiome-targeted therapies.
Associate Professor
Dr. Donia received his B.Sc in Pharmacy from the Faculty of Pharmacy, Suez Canal University, Egypt in 2004. He moved to the US in 2005 to study for his Ph.D. at the Medicinal Chemistry Department, School of Pharmacy, University of Utah. He worked in Dr. Eric Schmidt’s laboratory where he studied the chemistry and biology of small molecules produced by bacterial symbionts of marine animals. He used chemical, microbiological, and metagenomic techniques to study the role of small molecules in mediating microbe-host and microbe-microbe interactions in marine invertebrates. In 2010, he joined Dr. Michael Fischbach’s laboratory at the Department of Bioengineering and Therapeutic Sciences at the University of California, San Francisco. There, he studied small molecules produced by members of the human microbiome and their role in mediating microbe-host and microbe-microbe interactions in humans. In particular, he focused on antibiotics produced by human pathogens and commensals, and their role in shaping the composition and dynamics of the human vaginal and oral microbiota. Dr. Donia is a recipient of the NIH Director’s New Innovator Award, the Kenneth Rainin Foundation Innovation and Breakthrough Awards, and is named a Pew Biomedical Scholar. Dr. Donia is a member of the Scientific Advisory Board for Deepbiome Therapeutics.








wm6951[at]princeton.edu
ampe[at]princeton.edu
I received my B.S. in Biomedical Engineering from Washington University in St. Louis. Then, I completed my Ph.D. in Chemical and Biological Engineering at Princeton University studying the collective growth and death behavior of structured bacterial populations. In the Donia Lab, I study microbiome composition and interference in antibiotic treatment.
aw9423[at]princeton.edu
I earned a bachelor’s degree with majors in Biological Sciences and Psychology with a minor in Chemistry from Rutgers University. I completed a Ph.D. in Pharmaceutical Science at Rutgers’ Ernest Mario School of Pharmacy with a focus on pharmacokinetic and pharmacodynamic modeling. My dissertation examined how obesity alters drug exposure and response across a broad range of medications. In the Donia Lab, I study gut microbiome–mediated drug metabolism and investigate how microbiome-host interactions influence cancer progression and metabolic disorders. I value collaborative research and continual learning. Outside the lab, I enjoy exploring new cuisines, spirited debate, staying active, and fishing.
I grew up in New Hampshire and received my BA in Biology and History from Dartmouth College. I am broadly interested in applying machine learning methods to study the microbiome. Beyond the lab, I enjoy running, going to the gym, reading, and playing the guitar.
I grew up in South Jersey and earned my BS from Stetson University. Before starting graduate school, I completed a Post-Bacc at Cold Spring Harbor Laboratory, where I studied the intersection of cancer, diet, and the microbiome. I have a broad interest in endosymbionts, microbe-host interactions, and the role of the microbiome in drug metabolism. When I’m away from the lab, I enjoy pottery, spending time outdoors, or relaxing on the couch with a great book or show!

Name | Position in lab | Current |
| Ruojun Wang | Postdoctoral Researcher | Clinical Operations |
| Sarah Lekaj | Princeton Undergraduate ’25 | |
| Zoe Berman | Princeton Undergraduate ’25 | |
| Seema Chatterjee | Lab Manager/Technician | |
| Christina Kim | MA Graduate Student *25 | |
| Jongbeom Park | PhD Graduate Student *24 | Postdoc, University of Washington |
| Isabella Swartz | Princeton Undergraduate ’24 | |
| Katherine Carroll | Princeton Undergraduate ’24 | |
| Klara Thiele | Princeton Undergraduate ’24 | |
| Jeffrey Lee | PhD Graduate Student *23 | |
| Julia Garaffa | Princeton Undergraduate ’23 | Fulbright, Czech Republic |
| Ye Chen | Princeton Undergraduate ’23 | Graduate Student, University of Michigan |
| Jaime Lopez | PhD Graduate Student *22 | Postdoc, Stanford University |
| Mary Davis | Princeton Undergraduate ’22 | |
| Christian Hernandez | Princeton Undergraduate ’22 | PhD Student at UF Scripps Biomedical Research |
| Anna Schmedel | Faculty Assistant | |
| Shuo Wang | PhD Graduate Student *22 | Machine Learning Engineer, Apple |
| Amira Mira | Postdoctoral Research Fellow | Assistant Professor, Mansoura University, Egypt |
| Janie Kim | Princeton Undergraduate ’21 | Graduate Student, Stanford University |
| Nisha Chandra | Princeton Undergraduate ’21 | |
| Jared Balaich | PhD Graduate Student *20 | |
| Bahar Javdan | MD-PhD Graduate Student *20 | |
| Amir Erez | Postdoctoral Researcher | |
| Dani Peters | Princeton Undergraduate ’20 | |
| Francine Camacho | PhD Graduate Student *19 | Viome |
| Yuki Sugimoto | Postdoctoral Researcher | |
| Zhiyuan Li | Postdoctoral Researcher | Assistant Professor, Peking University |
| David Romero | Undergraduate SURP Intern | Undergrad, California State University, Northridge (CSUN) |
| Evan Zhao | Visiting Postdoctoral Researcher | Postdoc, Massachusetts Institute of Technology |
| Lucy Williamson | Princeton Undergraduate ’19 | |
| Pranatchareeya Chankhamjon | Postdoctoral Researcher | Scientist, VL55, Flagship Pioneering |
| Maria Diarey Tianero | Postdoctoral Researcher | Senior Scientist (Program Lead), Lodo Therapeutics |
| Mohamed Farag | Visiting Associate Professor | Associate Professor, Cairo University |
| Allison Chang | Princeton Undergraduate ’18 | Masters Student, Computer Science, Princeton University |
| Diana Chin | Princeton Undergraduate ’18 | Post-bac, Yu lab, Boston Children’s Hospital |
| Anna Posfai | Postdoctoral Researcher | Postdoc, Kinney lab, Cold Spring Harbor Laboratory |
| Tanya Tafolla | Summer Undergraduate Intern | Graduate Student, University of California, Merced |
| Jindong Zan | Postdoctoral Researcher | Staff Scientist, Becton Dickinson |
| Arman Odabas | Princeton Undergraduate ’17 | Med Student, Washington University in St. Louis |
| Audrey Abend | Princeton Undergraduate ’17 | Clinical Research Associate II, OpenBiome |
| Raphaella Hull | Oxford-Princeton Exchange Student | Graduate Student, University of Cambridge |
| Phoebe Huang | Princeton Undergraduate ’16 | Med Student, University of Rochester |
| Christian Shema Mugisha | Summer Undergraduate Intern | Graduate Student, Washington University in St. Louis |
 Mohamed S. Abou Donia
Mohamed S. Abou Donia Jie Liu
Jie Liu Paola Estrada
Paola Estrada Moamen Elmassry
Moamen Elmassry Laura Ióca
Laura Ióca Luis Jesus Linares-Otoya
Luis Jesus Linares-Otoya Sunghoon Hwang
Sunghoon Hwang Bhuwan Khatri Chhetri
Bhuwan Khatri Chhetri Wessam Mohamed
Wessam MohamedI received my B.S. in Biomedical Engineering from Washington University in St. Louis. Then, I completed my Ph.D. in Chemical and Biological Engineering at Princeton University studying the collective growth and death behavior of structured bacterial populations. In the Donia Lab, I study microbiome composition and interference in antibiotic treatment.
 Andrew Wassef
Andrew Wassef Catherine Day
Catherine Dayxc1590[at]princeton.edu
I received my B.S. in Chemical Engineering from UW-Madison. I am broadly interested in human health care and biotech. In the Donia Lab, I will be using both computational and experimental approach to study human gut microbiome. Outside of lab, I enjoy city explorations, hiking and occasionally skiing.
gcarver[at]princeton.edu
I grew up in NYC and received my B.S. in Biological Sciences from Cornell University, where I studied plant pathology. Before joining the Donia Lab, I spent two years working at the Broad Institute developing tools for genetic code expansion. Currently, my work focuses on ecologically and functionally relevant host-microbe interactions in marine symbioses. Outside of research I enjoy gardening and fermenting foods.
I grew up in the DC area and attended Dartmouth College in New Hampshire, where I studied biology and anthropology. My interests span the realm of microbiology and human health, including biofilm maintenance, the microbiome, and equity in healthcare. I’m interested in drug metabolism by the gut microbiome. When not at the bench, I enjoy embroidery, being outdoors, and spending time with friends.
I’m a New Jersey Native, having both grown up here and gone to school at Rutgers University-New Brunswick. Before Joining the Donia lab, I did 2 years of medical school at Robert Wood Johnson Medical School through the MD/PhD program. Currently, I’m using a multi-omics approach to investigate host-microbe interactions in the gut microbiome. Outside of the lab, I love coffee, watching formula 1 races, and riding my bike!
I got my B.S. in Biology with a minor in Sociology and Anthropology from Fordham University in NYC. In the Donia Lab, I am interested in understanding how the microbiome affects cancer progression. My hobbies include drawing, gardening, and cooking up a feast for friends and family.
 Sam Neff
Sam Neff
Quantitative and Computational Biology Graduate Student, joint with Mona Singh
sam.neff[at]princeton.edu
 Germaine Smart-Marshall
Germaine Smart-Marshall
Molecular Biology Graduate Student
gs0976[at]princeton.edu
 Noor Mohamed
Noor Mohamed Jillian Gullo
Jillian GulloFaculty Assistant
jg8071[at]princeton.edu
| Name | Position in lab | Current |
| Ruojun Wang | Postdoctoral Researcher | Clinical Operations |
| Sarah Lekaj | Princeton Undergraduate ’25 | |
| Zoe Berman | Princeton Undergraduate ’25 | |
| Seema Chatterjee | Lab Manager/Technician | |
| Christina Kim | MA Graduate Student *25 | |
| Jongbeom Park | PhD Graduate Student *24 | Postdoc, University of Washington |
| Isabella Swartz | Princeton Undergraduate ’24 | |
| Katherine Carroll | Princeton Undergraduate ’24 | |
| Klara Thiele | Princeton Undergraduate ’24 | |
| Jeffrey Lee | PhD Graduate Student *23 | |
| Julia Garaffa | Princeton Undergraduate ’23 | Fulbright, Czech Republic |
| Ye Chen | Princeton Undergraduate ’23 | Graduate Student, University of Michigan |
| Jaime Lopez | PhD Graduate Student *22 | Postdoc, Stanford University |
| Mary Davis | Princeton Undergraduate ’22 | |
| Christian Hernandez | Princeton Undergraduate ’22 | PhD Student at UF Scripps Biomedical Research |
| Anna Schmedel | Faculty Assistant | |
| Shuo Wang | PhD Graduate Student *22 | Machine Learning Engineer, Apple |
| Amira Mira | Postdoctoral Research Fellow | Assistant Professor, Mansoura University, Egypt |
| Janie Kim | Princeton Undergraduate ’21 | Graduate Student, Stanford University |
| Nisha Chandra | Princeton Undergraduate ’21 | |
| Jared Balaich | PhD Graduate Student *20 | |
| Bahar Javdan | MD-PhD Graduate Student *20 | |
| Amir Erez | Postdoctoral Researcher | |
| Dani Peters | Princeton Undergraduate ’20 | |
| Francine Camacho | PhD Graduate Student *19 | Viome |
| Yuki Sugimoto | Postdoctoral Researcher | |
| Zhiyuan Li | Postdoctoral Researcher | Assistant Professor, Peking University |
| David Romero | Undergraduate SURP Intern | Undergrad, California State University, Northridge (CSUN) |
| Evan Zhao | Visiting Postdoctoral Researcher | Postdoc, Massachusetts Institute of Technology |
| Lucy Williamson | Princeton Undergraduate ’19 | |
| Pranatchareeya Chankhamjon | Postdoctoral Researcher | Scientist, VL55, Flagship Pioneering |
| Maria Diarey Tianero | Postdoctoral Researcher | Senior Scientist (Program Lead), Lodo Therapeutics |
| Mohamed Farag | Visiting Associate Professor | Associate Professor, Cairo University |
| Allison Chang | Princeton Undergraduate ’18 | Masters Student, Computer Science, Princeton University |
| Diana Chin | Princeton Undergraduate ’18 | Post-bac, Yu lab, Boston Children’s Hospital |
| Anna Posfai | Postdoctoral Researcher | Postdoc, Kinney lab, Cold Spring Harbor Laboratory |
| Tanya Tafolla | Summer Undergraduate Intern | Graduate Student, University of California, Merced |
| Jindong Zan | Postdoctoral Researcher | Staff Scientist, Becton Dickinson |
| Arman Odabas | Princeton Undergraduate ’17 | Med Student, Washington University in St. Louis |
| Audrey Abend | Princeton Undergraduate ’17 | Clinical Research Associate II, OpenBiome |
| Raphaella Hull | Oxford-Princeton Exchange Student | Graduate Student, University of Cambridge |
| Phoebe Huang | Princeton Undergraduate ’16 | Med Student, University of Rochester |
| Christian Shema Mugisha | Summer Undergraduate Intern | Graduate Student, Washington University in St. Louis |
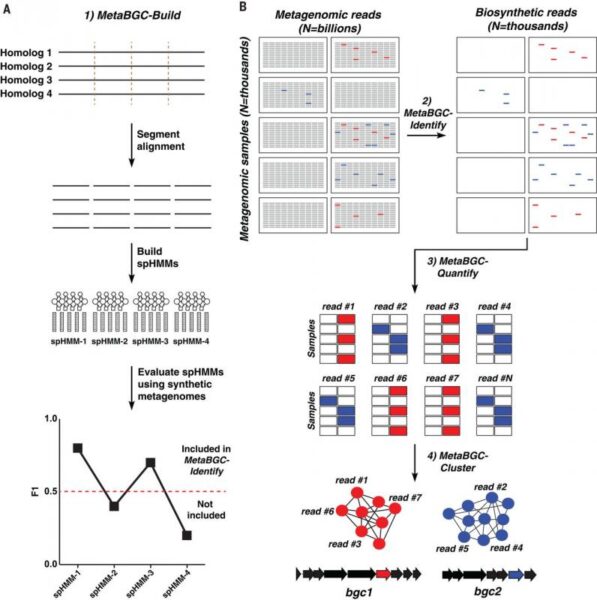
MetaBGC is a computational algorithm that discovers small-molecule BGCs directly in complex metagenomic sequencing data of the human microbiome.
To download MetaBGC, please visit our GitHub.
For more information about MetaBGC, please refer to our paper: Sugimoto, Y.*, Camacho, F.R.*, et al. (2019) “A metagenomic strategy for harnessing the chemical repertoire of the human microbiome”. Science. 366 (6471): eaax9176.
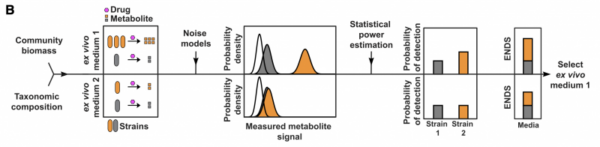
ENDS is a corrected richness metric where the contribution of each ASV is weighed by the probability that its metabolite can be detected while considering total biomass.
For more information on ENDS, please visit our GitHub and refer to our paper: Javdan, B.*, Lopez, J.*, et al. (2020). “Personalized Mapping of Drug Metabolism by the Human Gut Microbiome”. Cell. 7 (1661-1679): e22.
Jillian Gullo
217 Schultz Laboratory
jg8071[at]princeton.edu
(609)-258-5028